The topic of methodology in genomic EBVs was also very prominent at this conference. Already in the first plenary session, Mario Calus pointed out that this year, for the first time, more papers were submitted on genomic prediction than on GWAS and QTL prediction (Figure 1).
On the one hand, there were sessions that were specifically dedicated to the topic of “Methods and Tools”. The topic “Genomic Selection”, in particular single–step breeding value estimation, was also repeatedly addressed in additional sessions.
As a first example, Daniela Lourenco from the University of Georgia presented the further development of the BLUPF90 programs. This well-known program package was fundamentally revised. It can be used mainly for variance component estimation and traditional and genomic breeding value estimation. However, it can also be used to perform many upstream and downstream processes that are important for the above methods.
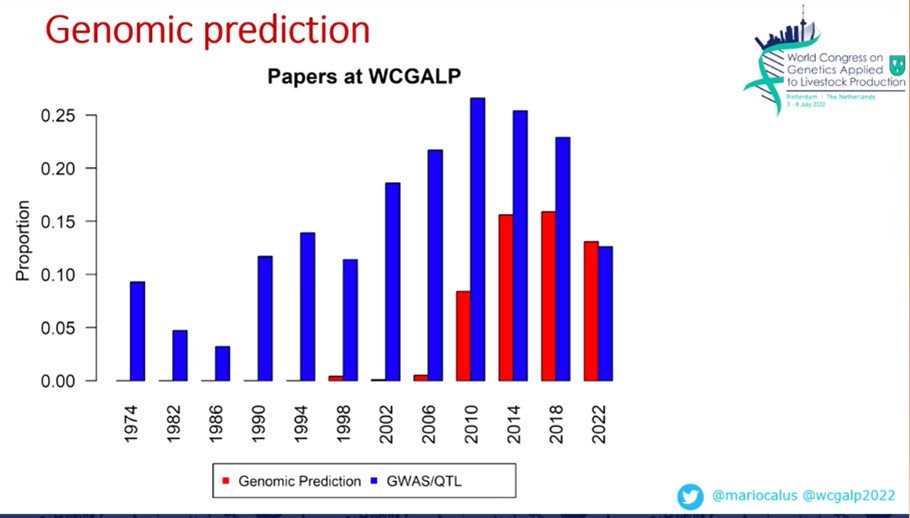
Figure 1: M.Calus WCGALP 2022 Number of submitted papers on GWAS/QTL (blue) and Genomic Prediction (red).
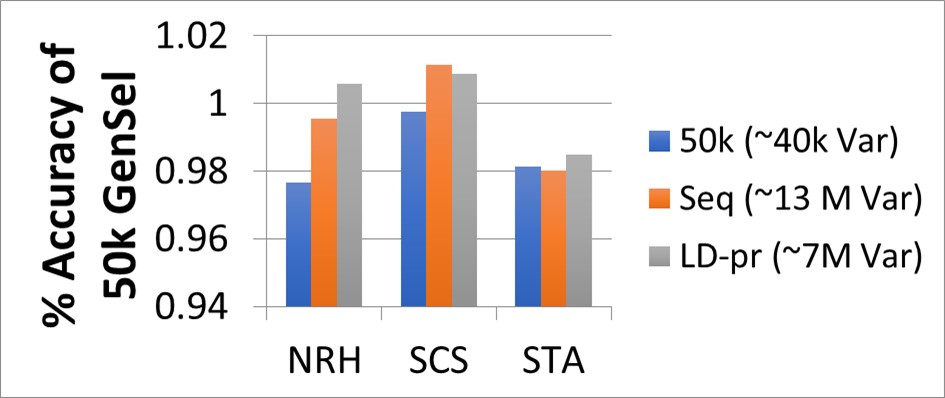
Figure 2: (Frischknecht et al. 2016, EAAP Belfast): Accuracy of genomic EBV with different marker densities compared to 50k density for the traits NRH: non-return rate bovine, SCS: somatic cell score and STA: sacral height.
In the session “Challenges – Use of whole genome sequence information” several contributions were shown on how to optimize the selection of markers for genomic selection. Several years ago, we argued that there is little gain in accuracy if all imputed sequence variants are considered. This basic finding was confirmed by several presentations in this session and new methods for the selection of an optimal marker set were presented.