The Plenary Session on Friday morning was entirely dedicated to genome-wide association studies. One of the world’s largest genome-wide association studies was presented here. This study incorporated size measurements and genotypes from nearly 5 million people. Loic Yengo of the University of Queensland in Brisbane, Australia presented this study. He highlighted interesting aspects of the inheritance of human size, but also pointed out the difficulties of such huge data sets. Especially in the human field, often only so-called “summary statistics”, rather than raw data, are shared and therefore some of the information is also lost. In the cattle area, some countries will probably soon have their own data sets of such sizes.
Genetic markers do not necessarily have to be typed via a commercially available chip. At Qualitas, a so-called “custom chip” has been in use for several years. The variants that are typed on this chip were selected by Qualitas geneticists in a project together with the University of Bern and designed and validated in collaboration with the laboratory and the chip manufacturer. Others are also following the path of a custom chip. For example, a custom chip was developed for the Schwarzbunte Niederungsrind, an old breed of cattle from Germany, taking population-specific markers into account.
Another approach to capture genotypes is the “low-pass-sequencing” methodology, a promising technology that has emerged in recent years and has since been further refined. In this technology, animals are sequenced genome-wide, but only with very low coverage. Missing markers are subsequently imputed with a reference panel. The goal is to achieve a much higher marker density than with chips.

Figure: Beef cattle (Image source: Karl Sulz)
Other presentations focused more on individual genomic regions. For example, Troy Rowan showed an alternative approach for identifying selection signatures: taking the year of birth of the animal as an approximation for the generation as the phenotype and doing a genome-wide association study. This was done for several North American beef cattle breeds and interesting results were obtained.
Aurelien Capitan, from INRAE in France, also expanded on the topic of lethal haplotypes in cattle in his presentation, so that haplotypes with incomplete penetrance can now also be identified. For this purpose, haplotypes were observed in different groups of animals. If the haplotypes occurred more often in groups where more animals died, this was an indication of such a haplotype with incomplete penetrance. The first variants have already been presented. The figure shows an example of how animals that are haplotype carriers survive.
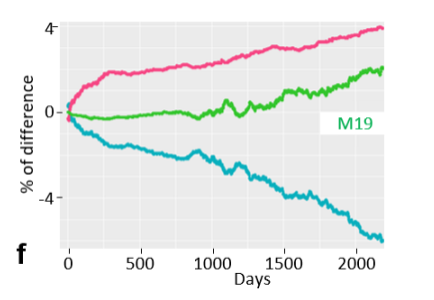
Figure: Difference in the departure rate of animals with different carrier statuses of the haplotype M19. Red: Died; Green: Slaughtered; Blue: Alive. In this haplotype, carriers have an increased risk of death throughout the entire breeding period. (Image source: Guintard, A. et al. 2022 Massive detection of cryptic recessive genetic defects in livestock mining millions of life trajectories., WCGALP).


