The Swiss Goat Breeders Association changes the technology for parentage control to SNP analysis. In the past weeks, the basics for SNP-based parentage control were established. SNP are selective changes in the genetic material. This makes them suitable for characterizing and clearly identifying different animals in a population.
Parentage control
Correct pedigrees are the basis for all genetic evaluations (breeding value estimation) and thus for breeding in general. In addition, correct pedigrees are also important for monitoring populations (inbreeding development).
Until now, the Swiss Goat Breeders’ Association (SZZV) used “microsatellites” to investigate parentage. These are now replaced by SNP analysis. As with the microsatellites, the results from the SNP analysis of an animal are compared with the results of its parents as part of the parentage control.
In the background are Mendel’s inheritance rules (see box at the end of the text). In the SNP-based parentage control, the SNPs are counted at which the Mendelian rules are violated when comparing animal – parent. The sum over all SNPs gives the result for the parentage control. A scheme with 10 SNPs as an example is shown in Figure 1.
Routinely 195 SNPs are used for this purpose in goats. In principle, a 100% match is the goal. However, since technical errors are also possible, also pedigrees with 1-2 errors on SNP level are accepted.
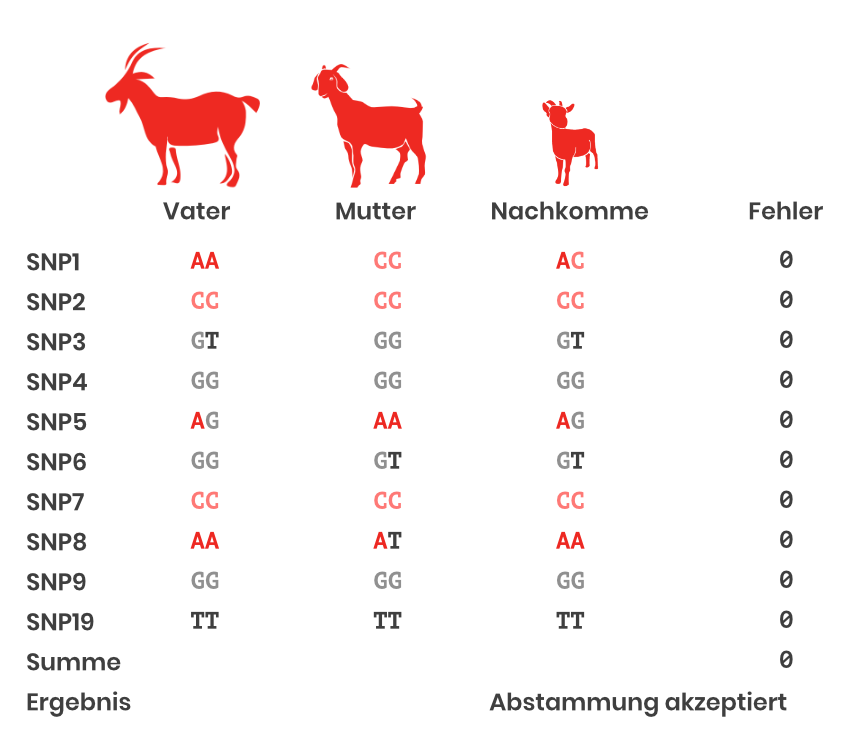
Figure 1: Parentage control with SNP (Design by Qualitas).
Benefit for the breeder
An essential difference to the previous system is that from now on the data from the examination are available to the SZZV. In the previous system, only the result from the pedigree check “pedigree accepted / rejected” was recorded and reported back to the breeder.
Nothing changes in the feedback to the animal owner, this is also limited in the future to the core statement “parentage accepted / rejected”. In the future, however, the 60,000 genotypes of each goat examined will be stored and will thus be available for future applications.
A first immediate usage for the breeder can be found in the search for possible parents. If the specified parentage is rejected, alternative parents are automatically searched for. The typing of almost 60,000 SNPs is therefore also suitable for searching for possible parents and for solving problems such as several male goats in a herd.
The search for possible parents is done in reverse to the parentage control. Among all genotyped animals, possible parents are searched and identified using all SNPs. This search is highly precise and reliable. This was not possible in the previous “microsatellite” system.
SNP – Single Nucleotide Polymorphism
Definition
The term SNP comes from the English language and means “single base exchange”. SNPs are found in the genetic material of goats. This comprises a total of about 3 billion bases, the so-called genetic letters. It is present in almost every cell of the organism. Because of its enormous size, it needs structure and organization. Two main basic structures are distinguished: chromosomes and double helix.
Chromosomes are subunits of the genetic material. The goat has 30 of them, and a goat inherits each of these chromosomes once from the father and once from the mother. The number of chromosomes is a characteristic of the species (Figure 2).
Each of these chromosomes consists of a so-called double helix. Two circulating strands are mutually twisted. Each strand is a sequence of genetic letters (A, C, G and T), which are chained together to form the strand.
A large part of these strands is identical between different animals, about 99%. The remaining 1% includes the SNP (Figure 3). In the case of a SNP, one animal carries e.g. an A and another animal a C at this position. Interesting for the animal breeder are the places where there are differences between the animals on the strands.
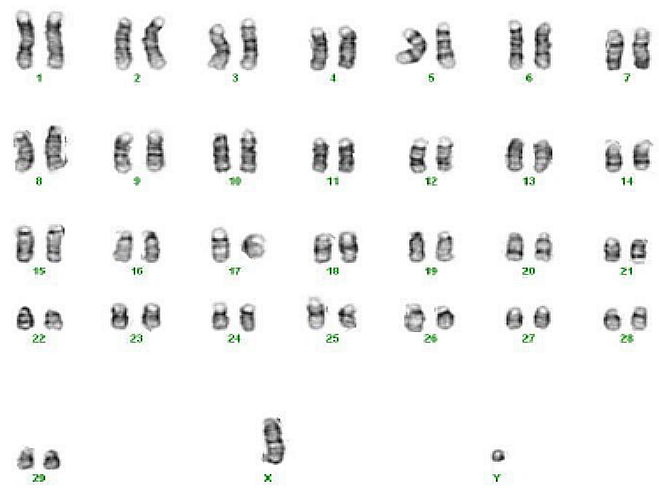
Figure 2: Chromosome set of a billy goat (karyotype).
Source: Hadj S Aoued and Mahipal Singh, Wikipedia.
Origin
SNPs arise spontaneously in the genome and often during the propagation of the genome. Depending on the position of the SNP and the cell in which it arises, the effect of a new SNP can be very different. SNPs that arise spontaneously, e.g. in a somatic cell, are not passed on to the offspring, but are restricted in their occurrence to the respective cell as well as its daughter cells from cell division. In contrast, SNPs that arise in the germ line, in the sperm or egg cells, are inherited over the generations.
Due to the behavior of breeders and the definition or separation of breeds, it is thus quite possible that a SNP is limited in its occurrence to one or a few breeds.
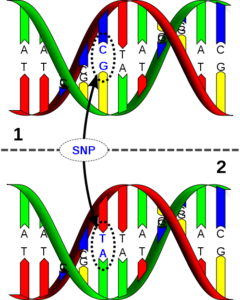
Figure 3: Single Base Exchange (SNP).
Source: Wikimedia.
Effect
Depending on the position of a SNP (inside or outside a gene), its functional effects are very different. Not every SNP necessarily has a (today known) function. Thus, the direct functionality of only a small fraction of all SNPs is known today. For the vast majority of SNPs, nothing or little of their effect is known even today.
SNPs with known effects include, for example, SNPs in color genes or milk protein genes. These SNPs lead to the expression of categorical traits such as protein type or color.
Detection of a SNP on a quantitative trait becomes even more difficult. Nevertheless, there is also a prime example here: the effect of a SNP from the gene called DGAT1 on the trait fat content. Animals with a C at this position in the genome have a significantly higher fat content in milk than animals with a T at the corresponding position.
Typing
Over the last decades, various technologies have been developed for the analysis of which letters are present in a SNP of an animal. This analysis is called typing.
The progress in this field has been enormous. Driven by the engine of basic research, SNP chips have now been available for farm animal species for more than 10 years. The best-known example of their use is probably genomic selection in cattle, where SNP chips have been essential since 2010.
In simplified terms, SNP chips are units (arrays) in which several thousand SNPs are examined simultaneously per animal in high throughput. Technological progress has also led to a massive reduction in costs. Consequently, typing today costs less than 0.1 Rappen per SNP. As a result, practical applications outside of research projects became possible in the first place.
Chips
As mentioned at the beginning, it must be assumed that there are around 30 million SNPs in the goat genome. This exceeds the possibilities of a chip, since a maximum of about 1 million SNPs can be analyzed with this technology. Thus, a selection must be made from the theoretically available SNP pool for chip typing.
A key property is the mutual location of the SNPs, since with a uniform distribution of the selected SNPs a network of markers is laid over the genome. This network is crucial for the following applications, including genomic selection.
For the widest possible range of applications of a chip, SNPs for parentage control should also be included. These must in fact fulfill an additional characteristic and be present in as many different breeds as possible, so that parentage control can be carried out with the same chip for all goat breeds.
Currently, the chip contains 195 SNPs for parentage control. Relevant additional tests e.g. kappa casein (milk protein) complete a chip. The chip, which will be used for parentage control, is an extension of the existing chip since 2014. It has been expanded especially in the area of additional tests. It was developed by an international consortium in which Swiss scientists from the Institute of Genetics at the University of Bern also contributed.

(Design by Qualitas)
Outlook
In the foreground is the introduction of the parentage control. However, as the chip contains a number of additional markers (kappa casein, DGAT1), their processing and publication will follow in the course of the year.
Through the international project SMARTER, in which genotypes of Swiss goats are also generated, the amount of data will increase rapidly, so that theoretically more advanced applications (genomic selection) will come within reach.
It is desirable if breeders make use of SNP typing as much as possible in their everyday life in the future, as this will quickly benefit goat breeding.


